EDIT: Thanks for comments, I created repository with full end-to-end reproducible code. You can find it here – https://github.com/dselivanov/kaggle-outbrain.
Recently I participated in Outbrain Click Prediction kaggle competition (and no, I won’t talk about crazy xgboost stacking and blending 🙂 ). Competition was interesting for me mainly because of 2 things:
- Organizers provided a lot of data – around 100gb. I like to solve problems efficiently, so initial main challenge for was to try to solve this on my laptop. In fact this is doable and I will descibe it (moreover, second place solution was done on laptop!).
- I hoped to earn “kaggle master”. For this I had to finish in top 11 teams. Unfortunately our team took 13th place. But this is also quite high position, and I want to share some ideas on how we managed to do this.
Task
In two words Outbrain is a platform which integrates to websites and shows some ads to visitors. Each block has 2-12 slots for ads. Train set consisted of ~17M events where customer clicked on one of the ads in such blocks. Goal was to rank ads accordingly and target was [Mean Average Precision @12 ](https://www.kaggle.com/wiki/MeanAveragePrecision). But I think as a proxy many of teams optimized logloss and evaluated AUC on cross-validation.
Data
As mentioned before, organizers provided several data files with relational structure. While all files were useful, some of them contribute more. We will be interesting in following files:
clicks_train.csv.zip,clicks_test.csv.zip– information about shown ads. Which advertisement identifiersad_idoutbrain presented for eachdisplay_idin ad block.events.csv.zip– provides information on thedisplay_idcontextpromoted_content.csv.zip– information about campaigns, advertisers and ids of the pages to which ad points.page_views.csv.zip– the log of users visiting documents
While most of the tables are small enough to fit in memory, the page views log (page_views.csv) is over 2 billion rows and 100GB uncompressed.
Libraries and tools
When you work with large amounts of data on a sinlge machine – your best friends should be UNIX command line utils and data.table package. Sparse matrices live in Matrix package. So our initial set will include:
set.seed(1)
library(data.table)
library(methods)
library(Matrix)
# like this sugar
library(magrittr)Also I want to introduce small wrapper on top of data.table::fread() which will read zipped files directly (without preliminary uznipping to disk):
fread_zip = function( file , ...) {
fn = basename(file)
# cut ".zip" suffix using substr
path = paste("unzip -p", file, substr(x = fn, 1, length(fn) - 4))
fread(path, ...)
}Also let’s parse promo and clicks_train files once and save them as serrialized R objects:
promo = fread_zip("~/projects/kaggle/outbrain/data/raw/promoted_content.csv.zip")
setnames(promo, 'document_id', 'promo_document_id')
saveRDS(promo, "~/projects/kaggle/outbrain/data/promo.rds", compress = FALSE)
clicks_train = fread_zip("~/projects/kaggle/outbrain/data/raw/clicks_train.csv.zip")
saveRDS(clicks_train, "~/projects/kaggle/outbrain/data/clicks_train.rds", compress = FALSE)Events
Lets start with events.csv and file which contains core information about display context.
events = fread_zip("~/projects/kaggle/outbrain/data/raw/events.csv.zip")
# several values in "platform" column has som bad values, so we need to remove these rows or convert to some value
events[ , platform := as.integer(platform)]
# I chose to convert them to most common value
events[is.na(platform), platform := 1L]Here is first trick. Since file is huge ( ~23M rows) and user ids uuid stored as strings, this will consume quite a lot of memory. Also such amount of string slows down R’s gc(). But we can hash these uuids into integers. Of course there will be some collisions, but if we will choose hashing range large enough, collision won’t affect result too much (I would say on this task they won’t hurt result at all). For hashing here I will use MurmurHash3 from text2vec. However this is not mandatory, you can create your own – for example take a look to digest package.
H_SIZE = 2**28
events[, uuid := text2vec:::hasher(uuid, H_SIZE)]Now we need to parse geo position. It is encoded in string and contais information about country, state and city. For the reasons described above we will also hash it. Reader should notice that hashing will be used a lot.
geo3 = strsplit(events$geo_location, ">", T) %>% lapply(function(x) x[1:3]) %>% simplify2array(higher = FALSE)
events[, geo_location := text2vec:::hasher(geo_location, H_SIZE)]
events[, country := text2vec:::hasher(geo3[1, ], H_SIZE)]
events[, state := text2vec:::hasher(geo3[2, ], H_SIZE)]
events[, dma := text2vec:::hasher(geo3[3, ], H_SIZE)]
rm(geo3)Mark events which are in train and test:
events[, train := display_id %in% unique(clicks_train$display_id)]Since we are working with time-series, it worth to check how organizers split data into train and test sets. So for each event we will calculate day from the first event:
events[, day := as.integer((timestamp / 1000) / 60 / 60 / 24) ]
saveRDS(events, '~/projects/kaggle/outbrain/data/events.rds', compress = FALSE)And check distribution:
library(ggplot2)
train_test_distr = events[, .N, keyby = .(day, train)]
ggplot(train_test_distr) + geom_bar(aes(x = day, y = N, fill = train), stat = 'identity')
train_test_distr[, N_share := N / sum(N), keyby = .(day)]
train_test_distr
ggplot(train_test_distr) +
geom_bar(aes(x = day, y = N_share, fill = train), stat = 'identity') +
scale_y_continuous(breaks=seq(0, 1, 0.05))
rm(train_test_distr)As you can see test set consists of events from days 13-14 and fraction (15%) of events from previous day. In real life it make no sense to predict “past”… However we will stick to distribution which is dictated by organizers data split.
set.seed(1)
events[, cv := TRUE]
# leave 11-12 days for validation as well as 15% of events in days 1-10
events[day <= 10, cv := sample(c(FALSE, TRUE), .N, prob = c(0.85, 0.15), replace = TRUE), by = day]
# sort by uuid - not imoprtant at this point. Why we are doing this will be explained below.
setkey(events, uuid)
# save events for future usage
#saveRDS(events, "~/projects/kaggle/outbrain/data/events2.rds", compress = FALSE)
# also for now let's remove test events, since we will work with local cross validation data
events = events[train == TRUE]Let’s check check how much memory we use at the moment:
tables()Around 2 gigs – any moderate laptop can hadle this. Even some smatrphones has 3-4 gb of ram 🙂
Now we are ready to build our first model.
Model matrix
As we see, all our feature are categorial variables. Lets try to create model matrix on 1/50 subset of data – we will take only uuids where uuid mod 50 = 0. One important note here – we partition data by uuid, but in theory we can also partition by display_id, ad_id, etc. Readers will see why we choose to partition by uuid in subsequent sections.
N_PART = 50
data_sample = events[uuid %% N_PART == 0]
# join with clicks_train to get information about clicks and shown ads
data_sample = clicks_train[data_sample, on = .(display_id = display_id)]
data_sample = promo[data_sample, on = .(ad_id = ad_id)]
object.size(data_sample) / 1e6Since we joined all parts, we can cosntruct feature matrix. But is also not obvious – how to encode categorial variables? Common approach is to calculate number of unique values for each factor and use 1-hot encoding. Other ideas? Hashing – we will define size of hashing space and project values of our categorial varibles in this space. By varying size of the hashing space we can control number of collisions. Let’s see how to implement this:
create_feature_matrix = function(dt, features, h_space_size) {
# 0-based indices
row_index = rep(0L:(nrow(dt) - 1L), length(features))
# note that here we adding `text2vec:::hasher(feature, h_space_size)` - hash offset for this feature
# this reduces number of collisons because. If we won't apply such scheme - identical values of
# different features will be hashed to same value
col_index = Map(function(feature) {
index = (text2vec:::hasher(feature, h_space_size) + dt[[feature]]) %% h_space_size
as.integer(index)
}, features) %>%
unlist(recursive = FALSE, use.names = FALSE)
m = sparseMatrix(i = row_index, j = col_index, x = 1,
dims = c(nrow(dt), h_space_size),
index1 = FALSE, giveCsparse = FALSE, check = F)
m
}
h_space_size = 2**24
feature_names = c("ad_id", "promo_document_id", "campaign_id", "advertiser_id", "document_id",
"platform", "geo_location", "country", "state", "dma")
X_train = create_feature_matrix(data_sample[cv == FALSE], feature_names, h_space_size)
y_train = as.numeric(data_sample[cv == FALSE, clicked])
data_sample_cv = data_sample[cv == TRUE]
X_cv = create_feature_matrix(data_sample[cv == TRUE], feature_names, h_space_size)
y_cv = as.numeric(data_sample[cv == TRUE, clicked])
str(X_train)
object.size(X_train)/1e6Note on memory consuption
And at this point it worth to think about how to solve such problem – model matrix with 1/50 of the dataset consumes ~ 200mb. Interpolating to full dataset – matrix will consume ~ 10gb. And this is already a problem for laptop with 16gb RAM. More interestingly it worth to point that at this moment model doesn’t contain any interactions between categorial variables, so prediction power is very limited! Moreover keep in mind that we have 100gb file of views… It becomes obvious that it will be hard to use batch algorithms which requires to kepp all data in RAM.
Note on algorithms and libraries
From my experience linear models work best on such wide high-dimensional sparse data sets. So xgboost (with tree booster) or RanomForest won’t work well. For such tasks I usually use glmnet package, it is also not ideal. For example on my laptop it can fit logistic regression with L1 penalty on matrix above (1/50 of data) for ~ 30 sec. But it consumes ~ 5gb(!) of RAM. Also not that here we check only 10 values of regularization parameter lambda (100 by default).
glmnet_model = glmnet::glmnet(x = X_train, y = y_train, family = 'binomial',
thresh = 1e-3, nlambda = 10, type.logistic = "modified.Newton")Other interesting packages I’m aware of: pirls, oem, bigLasso (does not support sparse matrices yet). But all of them are not well suited for us due to the size of the problem.
Online learning
It turns out that such large scale high dimensional problems can be efficiently solved in a streaming fashion with stochastic gradient descent. One notable example – vowpal wabbit.
Good thing about SGD is that it requires (O(1)) memory, but also vanilla SGD convergence rate heavily depends on learning rate. Fortunately it was well studied during recent years – it is used as optimization technique in all deep learning applications, used at internet scale ad prediction and so on.
It turns out that using different learning rates for each feature (adaptive learning rates) accelerate convergence drastically. Initial method was AdaGrad by Duchi et al. We will stick to article by McMahan which introduce state-of-the-art FTRL-Proximal algorithm for solving logistic regression with elastic-net penalty. It is proven to be useful in industry and recent CTR kaggle competitions.
FTRL proximal
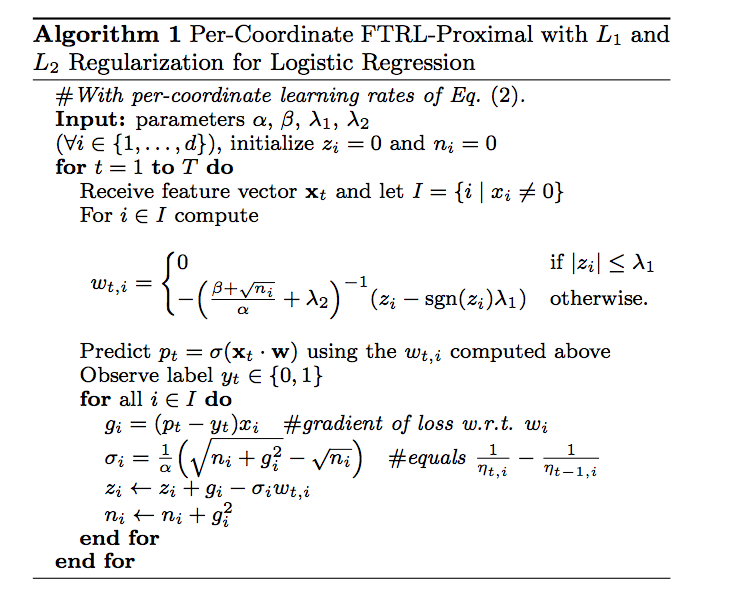
FTRLProximal
I won’t go into details about algorithm, reader can find details in papers above. I will focus on applying it to our problem. Honestrly speaking when I realized necessity of online algorithm, I started to search github for FTRL implementation. And for my surprise I found 3 implementations in R/C/Rcpp – 2 packages – rFTRLProximal, FTRLProximal and pure R implementation in FeatureHashing package (by the way this package allows to create hashed model matrices from data frames – all the stuff we did above manually).
First I tried packages and both of them had some issues:
rFTRLProximaldidn’t containt possibility of incremental learningFTRLProximalwas surprisingly very slow on my sparse data
Then I tried pure R solution from FeatureHashing which was also quite slow (which was not surprise. But it was faster than FTRLProximal!) After some profiling and refactoring I ended with quite fast pure R solution – you can find it on my gist.
And of course during competitions I made a lot of experiments, so didn’t want to wait long minutes while algorithm will converge. So I rewrote it using Rcpp and OpenMP, added dropout regularization and I’m happy to present FTRL package. Let me show it in action:
# first need to isntall it
devtools::install_github("dselivanov/FTRL")library(Matrix)
library(methods)library(FTRL)
# First of all need to convert matrix to row-sparse matrix
X_train_csr = as(X_train, "RsparseMatrix")
X_cv_csr = as(X_cv, "RsparseMatrix")
# set up model
ftrl = FTRL$new(alpha = 0.01, beta = 0.1, lambda = 20, l1_ratio = 1, dropout = 0)As can be noticed FTRL expose partial_fit() method (I follow scikit-learn convention in API). So model can be updated with new data. Or we can fit several epochs on same mini-batch. Let’s check timings with different number of threads:
# 1 thread
system.time(ftrl$partial_fit(X = X_train_csr, y = y_train, nthread = 1))
# 4 threads
system.time(ftrl$partial_fit(X = X_train_csr, y = y_train, nthread = 4))
# 4 threads + 4 hyperthreads
system.time(ftrl$partial_fit(X = X_train_csr, y = y_train, nthread = 8))Baseline
So now we are done with fast algorithm which can work on laptop. Let’s set up first baseline and put all together:
h_space_size = 2**24
feature_names = c("ad_id", "promo_document_id", "campaign_id", "advertiser_id", "document_id",
"platform", "geo_location", "country", "state", "dma")
X_cv = as(X_cv, "RsparseMatrix")
# regularization parameter lambda and learning rate alpha are important hyperparameters
# should be defined after via cross-validation and grid-search
ftrl = FTRL$new(alpha = 0.05, beta = 0.5, lambda = 1, l1_ratio = 1, dropout = 0)
# iterate through all examples
i = 1
for(i in 0:(N_PART - 1)) {
dt = events[uuid %% N_PART == i & cv == FALSE]
dt = clicks_train[dt, on = .(display_id = display_id)]
dt = promo[dt, on = .(ad_id = ad_id)]
# create model matrix for a given chunk and convert it to CSR format
X_train = create_feature_matrix(dt, feature_names, h_space_size) %>%
as("RsparseMatrix")
y_train = as.numeric(dt[, clicked])
# update model
ftrl$partial_fit(X = X_train, y = y_train, nthread = 8)
# check target metric. Alternatively can check map@12
# but in our case it was 100% correlated with AUC
if(i %% 5 == 0) {
train_auc = glmnet::auc(y_train, ftrl$predict(X_train))
p = ftrl$predict(X_cv)
dt_cv = data_sample_cv[, .(display_id, clicked, p = -p)]
# res[, p := -(p)]
setkey(dt_cv, display_id, p)
mean_map12 = dt_cv[ , .(map_12 = 1 / .Internal(which(clicked == 1))), by = display_id][['map_12']] %>%
mean %>% round(5)
cv_auc = glmnet::auc(y_cv, p)
message(sprintf("batch %d train_auc = %f, cv_auc = %f, map@12 = %f",i, train_auc, cv_auc, mean_map12))
}
}batch 0 train_auc = 0.724088, cv_auc = 0.705939, map@12 = 0.633280
batch 5 train_auc = 0.731734, cv_auc = 0.717310, map@12 = 0.640460
batch 10 train_auc = 0.733199, cv_auc = 0.720263, map@12 = 0.642470
batch 15 train_auc = 0.734564, cv_auc = 0.721715, map@12 = 0.642940
batch 20 train_auc = 0.735819, cv_auc = 0.722822, map@12 = 0.643520
batch 25 train_auc = 0.736287, cv_auc = 0.723580, map@12 = 0.643700
batch 30 train_auc = 0.735885, cv_auc = 0.723975, map@12 = 0.643940
batch 35 train_auc = 0.736175, cv_auc = 0.724659, map@12 = 0.644270
batch 40 train_auc = 0.737990, cv_auc = 0.725010, map@12 = 0.644170
batch 45 train_auc = 0.737410, cv_auc = 0.725422, map@12 = 0.645230
Along with leak it can bring us to ~150 position on leaderboard.
Baseline 2
One useful thing is to save chunks to disk for further model tuning, so will save model matrix for each mini-batch with wrapper of saveRDS function:
# by default saveRDS can save file with compression level = 6 - very slow
# or without compression - large files.
save_rds_compressed = function(x, file, compr_lvl = 1) {
con = gzfile(file, open = "wb", compression = compr_lvl)
saveRDS(x, file = con)
close.connection(con)
}Obvious thing to try is to add feature intercations and check how it will improve our model. In order to add interaction we need to modify our create_feature_matrix function. Interactions can be modelled as sum of corresponding underlying features because same combinations of features will be projected into the same values.
# now features will be a list of sets of features
# single values will correspont to single original features,
# while sets of features will correspond to interactions
# for example `features = list("platform", c("country", "campaign_id") )`
create_feature_matrix = function(dt, features, h_space_size) {
# 0-based indices
row_index = rep(0L:(nrow(dt) - 1L), length(features))
# note that here we adding `text2vec:::hasher(feature, h_space_size)` - hash offset for this feature
# this reduces number of collisons because. If we won't apply such scheme - identical values of
# different features will be hashed to same value
col_index = Map(function(fnames) {
# here we calculate offset for each feature
# hash name of feature to reduce number of collisions
# because for eample if we won't hash value of platform=1 will be hashed to the same as advertiser_id=1
offset = text2vec:::hasher(paste(fnames, collapse = "_"), h_space_size)
# calculate index = offest + sum(feature values)
index = (offset + Reduce(`+`, dt[, fnames, with = FALSE])) %% h_space_size
as.integer(index)
}, features) %>%
unlist(recursive = FALSE, use.names = FALSE)
m = sparseMatrix(i = row_index, j = col_index, x = 1,
dims = c(nrow(dt), h_space_size),
index1 = FALSE, giveCsparse = FALSE, check = F)
m
}And know we can create model matrix with some interactions:
interactions = c('promo_document_id', 'campaign_id', 'advertiser_id', 'document_id', 'platform', 'country', 'state') %>%
combn(2, simplify = F)
single_features = c('ad_id', 'campaign_id', 'advertiser_id', 'document_id', 'platform', 'geo_location', 'country', 'state', 'dma')
features_with_interactions = c(single_features, interactions)
h_space_size = 2**24
data_sample_cv = data_sample[cv == TRUE]
X_cv = create_feature_matrix(data_sample[cv == TRUE], features_with_interactions, h_space_size) %>%
as("RsparseMatrix")
y_cv = as.numeric(data_sample[cv == TRUE, clicked])
save_rds_compressed(list(x = X_cv, y = y_cv,
dt = data_sample_cv[, .(display_id, uuid, ad_id, promo_document_id, campaign_id, advertiser_id)]),
file = "~/projects/kaggle/outbrain/data/dt_cv_0.rds")As we will see process of each batch will take ~ 15 seconds. However on subsequent runs it will be much faster, because we will read model matrix for each minibatch from disk.
# will save mini-batches here
events_matrix_dir = "~/projects/kaggle/outbrain/data/events_matrix/"
dir.create(events_matrix_dir)
# iterate through all examples
for(i in 0:(N_PART - 1)) {
dt = events[uuid %% N_PART == 0 & cv == FALSE]
dt = clicks_train[dt, on = .(display_id = display_id)]
dt = promo[dt, on = .(ad_id = ad_id)]
setkey(dt, display_id, ad_id)
# create model matrix for a given chunk and convert it to CSR format
X_train = create_feature_matrix(dt, features_with_interactions, h_space_size) %>%
as("RsparseMatrix")
y_train = as.numeric(dt[, clicked])
# save file for further faster model tuning - we won't need to recalculate model matrix each time
# dt will be used in further steps to join with page views.
save_rds_compressed(list(x = X_train, y = y_train,
dt = dt[, .(uuid, ad_id, promo_document_id, campaign_id, advertiser_id)]),
file = sprintf("%s/%d.rds", events_matrix_dir, i))
message(sprintf("%s batch %d", Sys.time(), i))
}2017-02-04 18:55:58 batch 0
2017-02-04 18:58:46 batch 10
2017-02-04 19:01:29 batch 20
2017-02-04 19:04:14 batch 30
2017-02-04 19:06:56 batch 40
# regularization parameter lambda and learning rate alpha are important hyperparameters
# should be defined after via cross-validation and grid-search
ftrl = FTRL$new(alpha = 0.05, beta = 0.5, lambda = 1, l1_ratio = 1, dropout = 0)
for(i in 0:(N_PART - 1)) {
data = readRDS(sprintf("%s/%d.rds", events_matrix_dir, i))
# update model
ftrl$partial_fit(X = data$x, y = data$y, nthread = 8)
# check target metric. Alternatively can check map@12
# but in our case it was 100% correlated with AUC
if(i %% 5 == 0) {
train_auc = glmnet::auc(y_train, ftrl$predict(X_train))
p = ftrl$predict(X_cv)
dt_cv = data_sample_cv[, .(display_id, clicked, p = -p)]
setkey(dt_cv, display_id, p)
mean_map12 = dt_cv[ , .(map_12 = 1 / .Internal(which(clicked == 1))), by = display_id][['map_12']] %>%
mean %>% round(5)
cv_auc = glmnet::auc(y_cv, p)
message(sprintf("%s batch %d train_auc = %f, cv_auc = %f, map@12 = %f", Sys.time(), i, train_auc, cv_auc, mean_map12))
}
}2017-02-04 19:23:41 batch 0 train_auc = 0.720479, cv_auc = 0.710925, map@12 = 0.635120
2017-02-04 19:23:58 batch 5 train_auc = 0.736508, cv_auc = 0.725156, map@12 = 0.645800
2017-02-04 19:24:13 batch 10 train_auc = 0.740824, cv_auc = 0.729285, map@12 = 0.648530
2017-02-04 19:24:28 batch 15 train_auc = 0.743680, cv_auc = 0.731567, map@12 = 0.650900
2017-02-04 19:24:44 batch 20 train_auc = 0.745372, cv_auc = 0.733235, map@12 = 0.651650
2017-02-04 19:25:00 batch 25 train_auc = 0.746855, cv_auc = 0.734358, map@12 = 0.652510
2017-02-04 19:25:15 batch 30 train_auc = 0.748017, cv_auc = 0.734902, map@12 = 0.652590
2017-02-04 19:25:31 batch 35 train_auc = 0.748976, cv_auc = 0.736185, map@12 = 0.653550
2017-02-04 19:25:48 batch 40 train_auc = 0.749709, cv_auc = 0.736798, map@12 = 0.653320
2017-02-04 19:26:03 batch 45 train_auc = 0.750500, cv_auc = 0.737207, map@12 = 0.654440
Again, with leak (which add ~ 0.015) this will give score ~ 0.67 bring to ~ 90-100 position on leadderboard. Please, note time we spend for data preparation and model fitting – we were able to get into top 100 in 20-25 minutes on a laptop.
Page views
The most interesting file is views.csv.zip (100 gb of logs for customer visits!). I will describe on how to effectively incorporate information from it in next blog post. Stay tuned.